|
|
|
Mouse Genome
Databases
This server provides data extracted and compiled from
- The 2000-2001 Mouse Chromosome Committee Reports
- Release 15 of the MIT microsatellite map (Oct 1997)
- The recombinant inbred strain database of R.W. Elliott (1997) and R.
W. Williams (2001)
- Map Manager and text format chromosome maps (Apr 2001)
Most files are intended for you to download and use on your own
personal computer. Most files are available in generic text format or as
FileMaker Pro databases. All files can be used with Macintosh and Windows
operating systems.
Mouse Genome Databases
Note: To use FileMaker and Excel files listed below you may need to
configure your web browser to recognize the appropriate file type. You
will also need a copy of FileMaker Pro 3.0 or higher and Excel 98 or
higher. When you first open FileMaker Pro databases you will be greeted
with the message: "This file was not closed properly. FileMaker is now
performing a consistency check." This message is normal.
-
LXS genotype Excel file Updated, revised positions for 330 markers genotyped using a panel of 77 LXS strain.
-
MIT SNP DATABASE ONLINE: Search and sort the MIT Single
Nucleotide Polymorphism (SNP) database ONLINE. Fifty records are
displayed per page in the "list" format. These data from the
MIT-Whitehead SNP release of December 1999.
- INTEGRATED MIT-ROCHE SNP DATABASE in
EXCEL and
TEXT FORMATS (1-3 MB): We have merged the original
MIT SNPs with the new
Roche SNPs. The Excel file has been formatted to illustrate SNP
haplotypes and genetic contrasts. Both files are intended for
statistical analyses of SNPs and can be used to test a method outlined
in a paper by Andrew Grupe, Gary Peltz, and colleagues (Science 291:
1915-1918, 2001). The Excel file includes many useful equations and
formatting that will help in navigating through this large database and
in testing the "in silico" mapping method. Download the
Excel file to disk.
Contact Rob Williams (rwilliams@uthsc.edu) if you have trouble
downloading or interpreting these files. View an
example of the Excel spreadsheet near the Nnc1/Thra/Rara loci on Chr
11 (58 cM).
Our own analysis does not support the idea that the current set of
SNPs will be useful for discovery or confirmation of QTLs. The data
themselves are fascinating, but there are on average only two or three
distinct SNP haplotypes across any region of the genomes of standard
inbred strains. Furthermore, as indicated by Grupe et al., recombination
densities of SNP haplotypes are surprisingly low reducing the potential
resolution. These two characteristics of SNP maps negate their use to
map even Mendelian loci. The apparent success of Grupe et al. in using
this approach to confirm QTLs (including two of our own; eye weight and
retinal ganglion cell number) is a matter of ongoing debate. In any
case, you can look over the complete SNP genotypes files now to judge
for yourself.
-
Use of inbred strains for the study of individual differences in pain related phenotypes in the mouse:
Elissa J. Chesler's 2002 dissertation, discussing issues relevant to the integration of genomic and phenomic data from standard inbred strains
including genetic interactions with laboratory environmental conditions and
the use of various in silico inbred strain haplotype based mapping
algorithms for QTL analysis.
- SNP QTL MAPPER in
EXCEL format (572 KB, updated January 2002 by Elissa Chesler):
This Excel workbook implements the Grupe et al. mapping method and
outputs correlation plots. The main spreadsheet allows you to enter your
own strain data and compares them to haplotypes. Be very cautious and
skeptical when using this spreadsheet and the technique. Read all of the
caveates. This excel version of the method was develped by Elissa Chesler.
This updated version (Jan 2002) handles missing data. Download the
spreadsheet to disk.
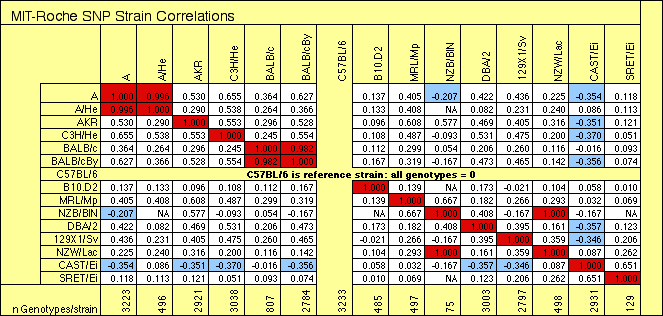
Correlation matrix for strains included
in the Integrated SNP files. This image is a clipping from the
Excel file. The two right-most strains, SPRET/Ei and CAST/Ei, are
different species and subspecies, respectively, and both need to be
treated with special care in any comparative SNP analysis. Some pairs of
substrains are > 98% identical.
-
MIT SNP Database DOWNLOAD the database in tab-delimited text format.
This file is suitable for manipulation in statistics and spreadsheet
programs (752 KB, Updated June 27, 2001). Data have been formatted in a
way that allows rapid acquisition of the new data from the
Roche Bioscience SNP database.
-
MIT SNP Database (DOWNLOAD the FileMaker 5 Version): This is
a reformatted version of the MIT Single Nucleotide Polymorphism (SNP)
database in FileMaker 5 format. You will need a copy of this application
to open the file (Mac and Windows; 992 KB. Updated July 13, 2001 by RW).
-
Gene Mapping and Map Manager Data Sets: Genetic maps of mouse
chromosomes. Now includes a 10th generation advanced intercross
consisting of 500 animals genetoyped at 340 markers. Lots of older files
on recombinant inbred strains.
-
The Portable Dictionary of the Mouse Genome, 21,039 loci,
17,912,832 bytes. Includes all 1997-98 Chromosome Committee Reports and
MIT Release 15.
-
FullDict.FMP.sit: The Portable Dictionary of the Mouse
Genome. This large FileMaker Pro 3.0/4.0 database has been compressed
with StuffIt. The Dictionary of the Mouse Genome contains data from the
1997-98 chromosome committee reports and MIT Whitehead SSLP databases
(Release 15). The Dictionary contains information for 21,039 loci.
Download this version if you have StuffIt or UnStuffIt. File size = 4846
KB. Updated March 19, 1998.
-
MIT Microsatellite Database ONLINE: A database of MIT
microsatellite loci in the mouse. Use this FileMaker Pro database with
OurPrimersDB below. MITDB is a subset of the Portable Dictionary of the
Mouse Genome. ONLINE. Updated July 12, 2001.
-
MIT Microsatellite Database: A database of MIT microsatellite
loci in the mouse. Use this FileMaker Pro database with OurPrimersDB
below. MITDB is a subset of the Portable Dictionary of the Mouse Genome.
File size = 3.0 MB. Updated March 19, 1998.
-
OurPrimersDB: A small database of primers. Download this database if
you are using numerous MIT primers to map genes in mice. This database
should be used in combination with the MITDB as one part of a relational
database. File size = 149 KB. Updated March 19, 1998.
-
Empty copy (clone) of the Portable Dictionary in FileMaker Pro 3.0
format. Download this file and import individual chromosome text
files from the table below into the database. File size = 231 KB.
Updated March 19, 1998.
Chromosome Text Files from the Dictionary
The table to the left lists data on gene loci for individual
chromosomes. Data types in these files are separated by tab characters,
and each gene or marker locus is separated by a carriage return. The first
record in each file lists the names of the variables. You can import the
tab-delimited text files into a word processor, but it will be hard to
read (adjusting the separation between tabs and creating a hanging indent
will help). The files are easy to handle with spreadsheet or database
programs (Excel, Filemaker Pro, etc.). Files are sorted alphabetically by
locus symbol. If you import data into a spreadsheet or database, you can
easily resort the loci by position (Map_ccr = map position from the
1997-98 Mouse Chromosome Committee Reports; Map_mit = map position from
the MIT microsatellite map).
To download one of these files, hold down the mouse button over the
link until a pop-up menu appears, then select the "Save this Link as..."
option in the menu. On a Macintosh, you can also just option-click the
link to save the file to disk. Another way of saving the file is to load
the file normally and just save it as text from within your web browser.
You can open the file that you have downloaded using a spreadsheet or
database program. If you import text files into Excel, be sure to import
it as a tab-delimied file (select Tab-delimited Options in the
Open menu).
The text files in this table were extracted from the Portable
Dictionary of the Mouse Genome. This dictionary includes data on 21,039
loci (genes and anonymous DNA segments). It contains the following types
of information:
- current symbols, old symbols, names, and aliases
- three sets of data on the positions of loci
- from the November 1993 GBASE Locus Map
- from the 1993 and 1994 Chromosome Committee Reports
- data from the April 1995 MIT SSLP database release
- sequence accession numbers associated with gene loci
- locations of homologous genes in human and 10 other mammalian
species
- recombinant inbred strain data
- strain distribution data of alleles
- probe and PCR primer data
- enzyme commission numbers
- phenotype codes
- references (in progress)
These files are generally smaller than 500k. They include data from
Release 15 of MIT microsatellite loci (Oct 97).
Updated by
Rob Williams and
Alex Williams.
|
|
|

 Databases
Databases Page maintained by R.Williams | A.Centeno. Last
updated:
Page maintained by R.Williams | A.Centeno. Last
updated:
